What is our vision?
Within the SMARTDIAGNOS project we will develop innovative diagnostic equipment enabling fast identification of the pathogens causing sepsis and antimicrobial resistance profiles. We envisage pathogen detection directly in whole blood with minimal sample handling. Additionally, we might add particular settings where our developments could be superior (slow growing pathogens, pathogens with particular culture requirements, etc.).
Why are current solutions for sepsis laboratory diagnostics not good enough?
Today, routine clinical applications for detection of pathogens in sepsis patients are too slow to give a real impact on treatment options. The current gold standard, blood culture followed by species identification and resistance detection on plate-cultured bacterial isolates, will take approximately 2-5 days before final results are achieved. For negative blood cultures the response time is even longer. New techniques are used to speed up the time of this method, such as species identification by Maldi-Tof MS, fluorescent in situ hybridization (FISH) and automated resistance detection. All these improvements of the routine method reduce the time to result, but cultured isolates are still needed, which takes time.
Our solution
The two new solutions for diagnostics will, in combination, provide early and accurate pathogen detection for an improved treatment. This will influence clinical decision making and drastically reduce morbidity and mortality and long-term side effects by enabling earlier targeted antimicrobial therapy. Fast and appropriate treatment for sepsis is important since “Every Minute Counts”!
Sepsis and antimicrobial therapy
Immediate antibiotic therapy is recommended by the International Guidelines for the Management of Severe Sepsis and Septic Shock, and early adequate antimicrobial therapy was shown to significantly improve the survival of septic patients. The guidelines also mandate pathogen identification by microbiological methods (blood culture) prior to the initiation of antibiotic therapy wherever possible. As blood culture is associated with a time-to-result of 48 - 72 hours, broad-spectrum antibiotics are used initially to ensure coverage of potential pathogens, which, however, is costly, associated with the risk of overtreatment, potentially toxic, and can result in the selection of multidrug-resistant bacteria, representing a significant public health threat. Blood culture may fail to detect slow-growing or intracellular pathogens, such as Rickettsia spp., Bartonella spp., Coxiella spp., Mycoplasma spp., or Chlamydia spp., or may remain negative due to concurrent antibiotic treatment even when bacterial or fungal infections are strongly suspected.
New diagnostic tools for sepsis
Therefore, there is a currently unmet medical need for reliable, fast, sensitive, and culture-independent diagnostic tools for pathogen detection directly from patient specimens. One approach in this respect is the use of molecular diagnostics based on polymerase chain reaction (PCR), providing advantages such as short time-to-result of 4 - 7 hours as compared to 48 - 72 hours for blood culture, small sample volume, applicability with concurrent antibiotic treatment, and absence of time consuming and potentially pre-selecting culture steps. While current PCR-based systems appear to be less sensitive and less specific when compared to blood culture, they may be superior to blood culture in certain settings, such as for the quantification of intracellular pathogens, for pathogen detection after initiation of antibiotic treatment, or in chronic, recurrent, culture-negative infections. The inclusion of biomarkers, such as acute phase proteins, cytokines, or chemokines to complement molecular diagnostics may further improve pathogen detection.
Molecular diagnostics
Several nucleic acid based technologies, NAATs, are commercially available today for identifying pathogens directly in positive blood culture bottles. Diagnostic tests for detection of pathogens directly from whole blood are also in the market, but these methods suffer from low diagnostic sensitivity due to small amounts of blood used for the test and a too narrow panel of pathogens/resistance markers included. Organisms are known to be present in very low counts per mL of blood, which makes detection difficult. So far, no molecular method have transformed into a new gold standard replacing traditional blood cultures for diagnosis of sepsis, due to complex handling and lengthy procedures, as well as low clinical relevance of results.
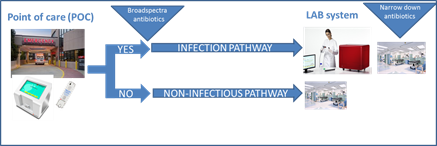
POC and LAB systems
Our proposed solution for this is the use of a point-of-care (POC) system answering the question of the presence of infection or non-infectious SIRS (systemic inflammatory response syndrome). In this system, tests for specific sepsis-related miRNA produced as a result of bacterial infection and broad range bacteria/yeast detection will be performed. The POC system will give a fast response, within 1 h, to enable an evidence-based initial diagnosis to start antimicrobial treatment when necessary. A negative test for infection can be used to rule out pathogens as the cause of the symptoms. If the result is “infection present”, the sample will be further tested by the laboratory (LAB) system which includes a broad panel of pathogens and resistance markers tested on a larger volume of whole blood, 5-10 mL. The LAB system will give the broader picture and confirm, or adjust, the initial diagnosis. In this project, technical innovation are of major importance for the success. In the last year of the project, the performance of instrument prototypes will be evaluated in clinical laboratory settings using real patient samples. A larger clinical validation needs to be performed after this project ends, prior to marketing.